The Solvent-Excluded Surface (SES) is an essential representation of molecules which is massively used in molecular modeling and drug discovery since it represents the interacting surface between molecules. Based on its properties, it supports the visualization of both large scale shapes and details of molecules. While several methods targeted its computation, the ability to process large molecular structures to address the introduction of big complex analysis while leveraging the massively parallel architecture of GPUs has remained a challenge. This is mostly caused by the need for consequent memory allocation or by the complexity of the parallelization of its processing. In this paper, we leverage the last theoretical advances made for the depiction of the SES to provide fast analytical computation with low impact on memory. We show that our method is able to compute the complete surface while handling large molecular complexes with competitive computation time costs compared to previous works.
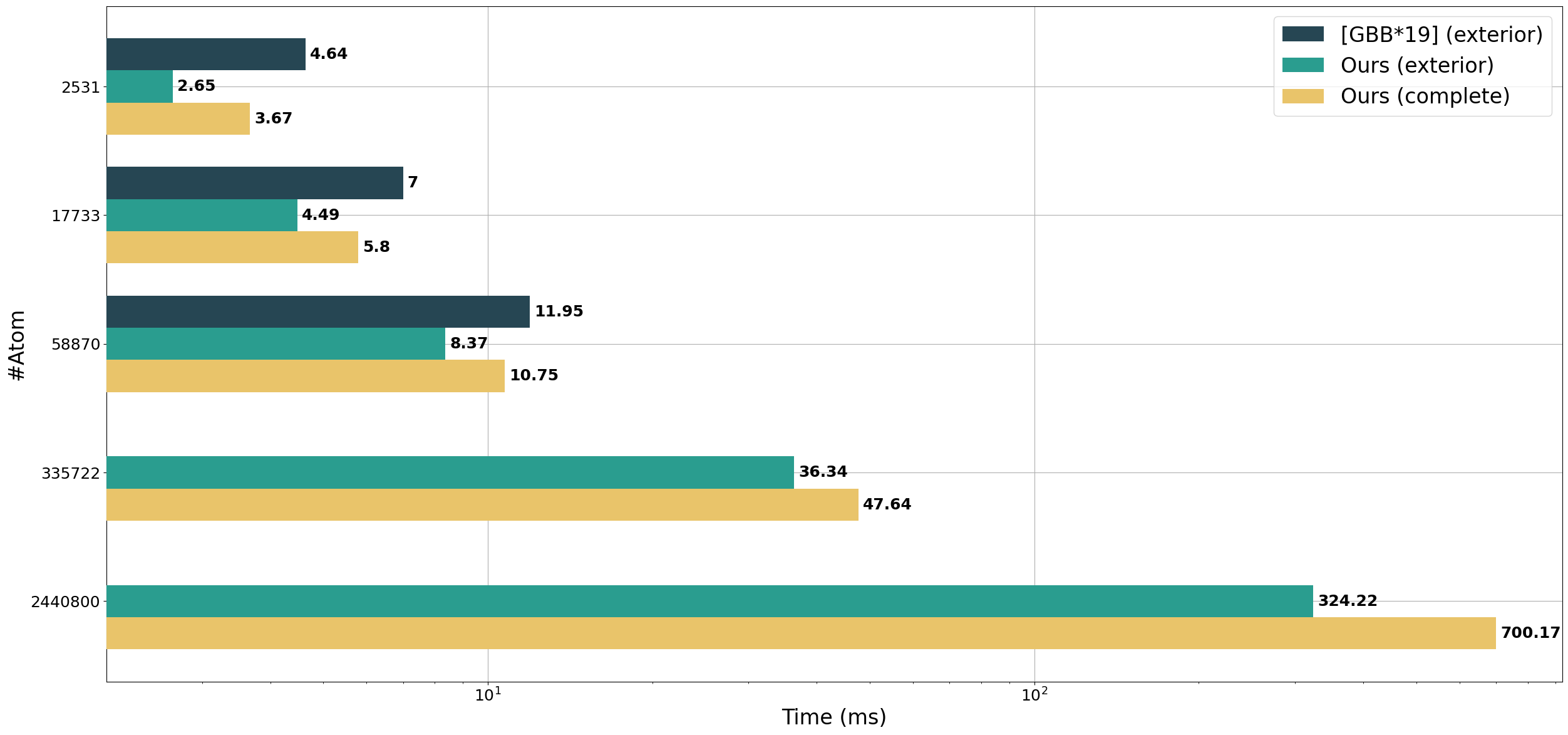
We compare our method to Megamol's implementation of GPU Contour-Buildup, the fastest method in our experiments. On the same GPU, our method is able to compute the surface of 30x larger protein without being restricted to the exterior surface.
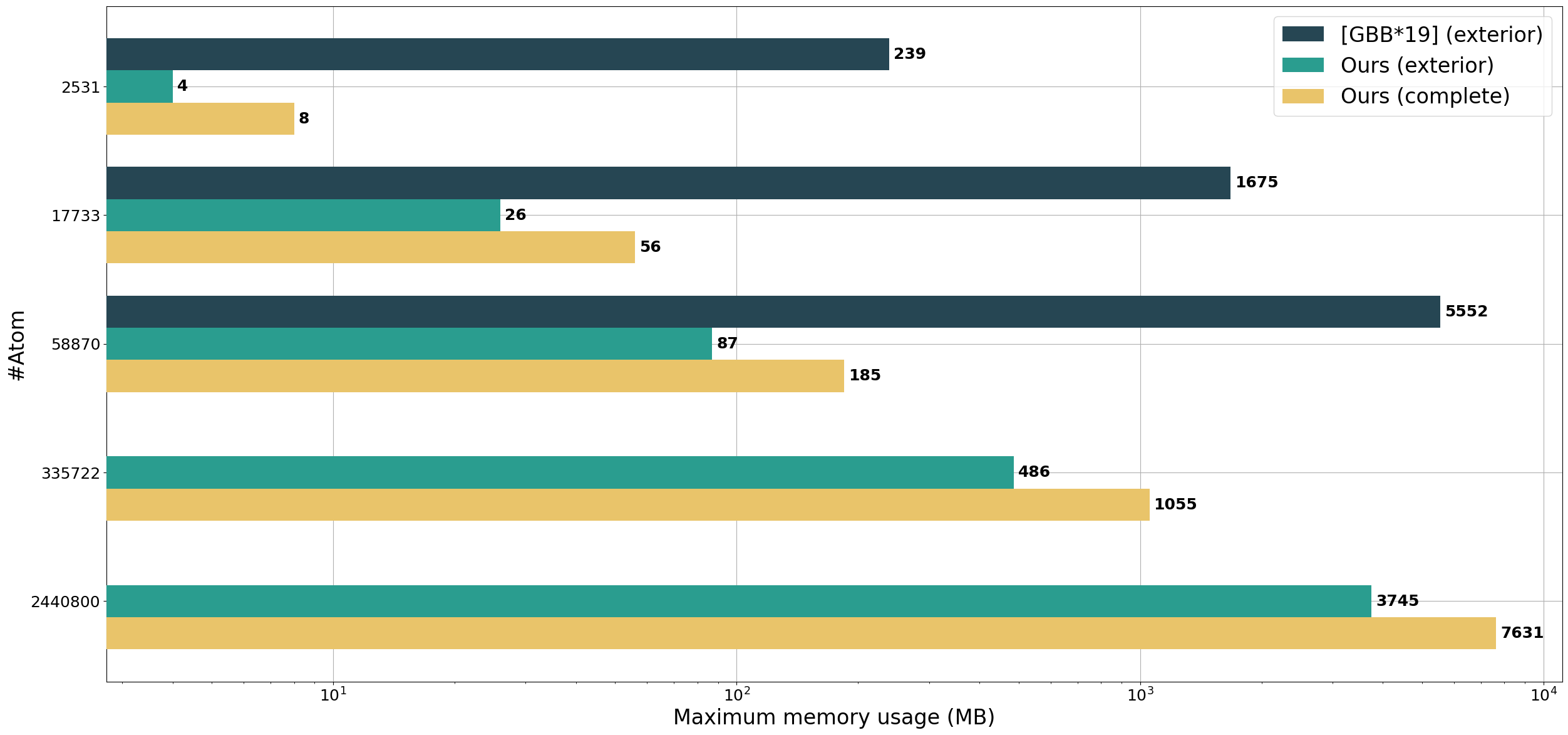
Regarding computation time, our methods perform similarly to GPU Contour-Buildup, which allows the fast computation of the complete surface of large proteins.
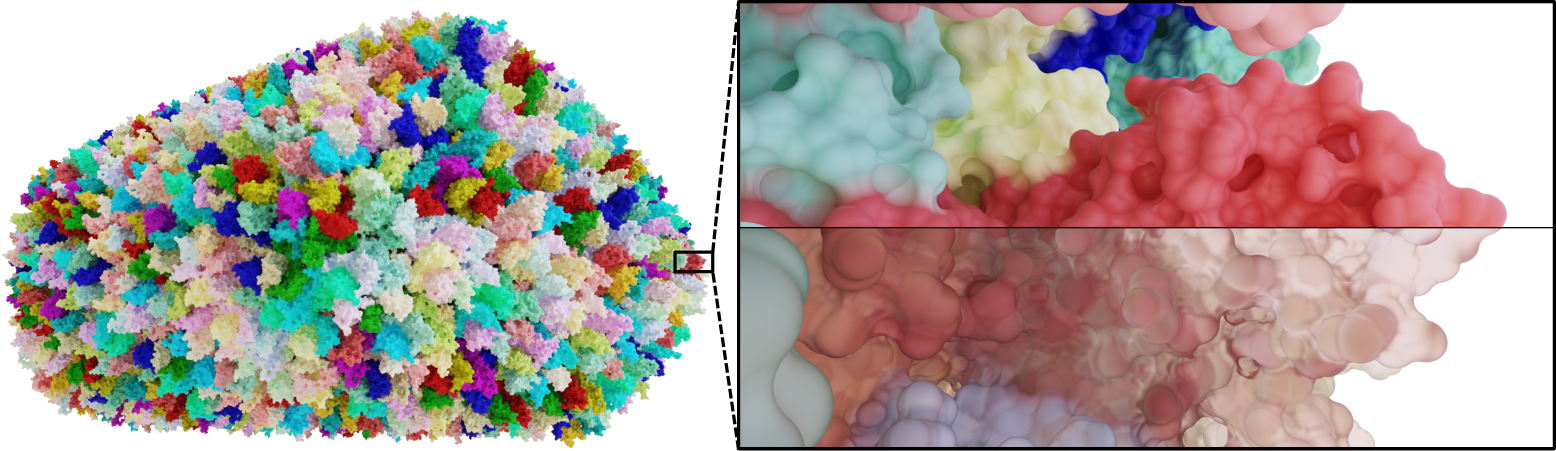
Thanks to reduced computation time and complete geometry computation, we can compute illustrations of molecular bodies with various atom counts and shading effects emphasizing the properties of the molecular surface.
Finally, our method can be used for interactive visualisation of proteins with per-frame updates.
Per-frame update with randomized geometry update. PDB ID: 1AON
@article{PlateauHolleville2024,
author={Plateau—Holleville, Cyprien and Maria, Maxime and Mérillou, Stéphane and Montes, Matthieu},
journal={IEEE Transactions on Visualization and Computer Graphics},
title={Efficient GPU computation of large protein Solvent-Excluded Surface},
year={2024},
volume={},
number={},
pages={1-12},
doi={10.1109/TVCG.2024.3380100}
}